Anthony Salvagno, a grad student in our lab recently described his encounter with the fear of scooping on our lab blog. In one of my responses to him in the comments, I state how I really don't think we're competing with the Wang lab, or any other lab, as far as I know at this moment. We are, of course, in competition with other labs, in terms of struggle to obtain a share of limited research funding. But I don't want our lab to get mired in direct competition and "race to publish first" that sometimes occurs. I'm not so worried about this, though, because I think we have a whole slew of important and novel ideas that we can pursue--much more than we have the manpower for. As Michael Nielson pointed out to me in the comments to his blog post, most scientists are in this boat: they have far more good research ideas than they can pursue. This is what makes intentional scooping a rare event in my mind -- it is only carried out by those paranoid, non-creative PIs whose fear of failure forces them to steal other's ideas and possibly crush younger scientists along the way. I think as Open Science (aka Science 2.0) takes over in the next decades, episodes of intentional scooping will become much, much easier to punish, due to the public track record of research progress and grant proposals available for all to see.
In this post, I want to elaborate on why I think our ideas are important but unique, and therefore not in direct competition with most of the leading labs in the single-molecule manipulation world today. The table below illustrates why I think we can lead a new era of experiments in an under-tapped area of single-molecule analysis--"single-molecule genetics." I am using a 2x2 matrix to analyze the research space. This is a technique I learned during the end of my graduate career when I interviewed and almost landed a job with Boston Consulting Group (BCG, a company I highly respected). The top row represents ensemble assays where the properties of many molecules are averaged together. The bottom row represents the single-molecule analysis research world. The left column represents experiments where the system being studied has been reconstituted from purified components. The right column represents experiments where the system being studied was intact, with only a few specific genes having been mutated or knocked-down*.
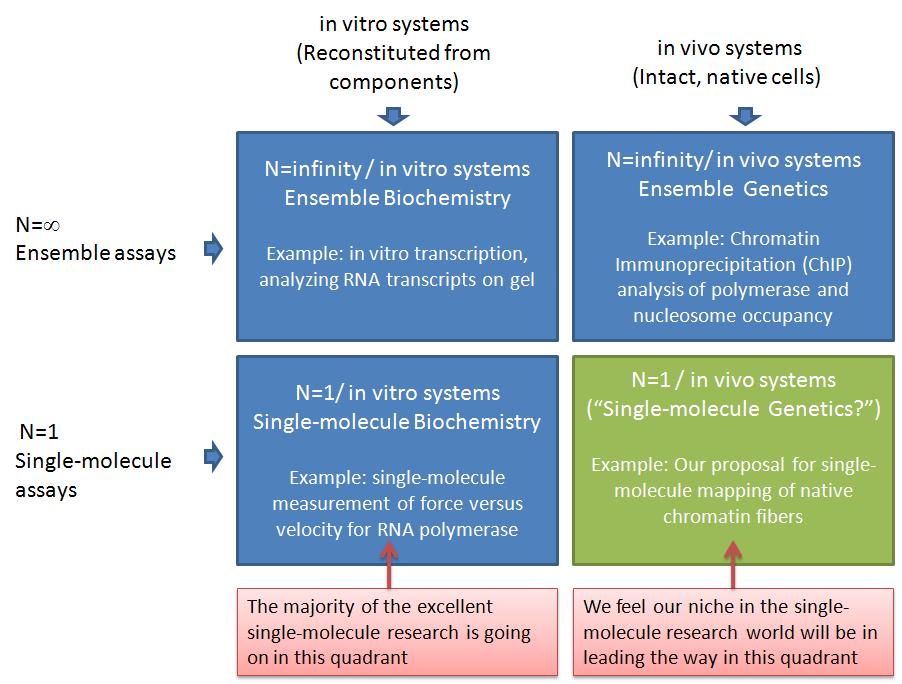
The top left quadrant represents what people refer to as biochemistry research, whereas the top right quadrant would be genetics**. The vast majority of the important discoveries in biology in the past half century would be in the top row (ensemble assays). Single-molecule assays in the bottom row have been very valuable, but are only now becoming more widely available. In my opinion, the vast majority of single-molecule assays have been in in the bottom left quadrant, which I am calling "single-molecule biochemistry" for convenience. For example, the Steve Block lab has made a number of breakthroughs in this quadrant. The Block Lab is a good example, because the research and papers produced by the students and postdocs in this lab over the past couple decades are of the highest quality and have played a key role in defining the single-molecule field. One of their areas of study is RNA Polymerase, where they have applied forces to recombinant E. coli RNA Polymerase during elongation in all sorts of manners: opposing elongation, assisting elongation, pulling on the RNA transcript, etc. Another main area of the block lab is the molecular motor kinesin. They have used optical tweezers to make many important discoveries about the mechanochemistry of this amazing enzyme. From these two areas combined (which still account for only part of the Block Lab research), they have something like 10,000 Science, Nature, and Cell papers. And as far as I know, all of that research has been done using dozens of different recombinant RNA Polymerase and kinesin heavy chain (KHC) motor proteins. So, as far as my matrix goes, they have made a huge impact on the bottom left quadrant -- "single molecule biochemsitry."
As I've said, I think the bottom right quadrant has so far been under-utilized. I am calling this quadrant "single-molecule genetics," to highlight what I see as the untapped power of combining proven single-molecule analyses with existing genetics techniques. When I say "untapped," I don't mean to imply that there aren't existing studies in this quadrant. I only mean that most of the research (and money) from single-molecule manipulation groups I am familiar with has been going into the SM biochemistry quadrant. This includes groups such as those of Steve Block, Carlos Bustamante, Evan Evans, Mark Williams, Jeff Gelles, Vincent Croquette, David Bensimon, Steve Kowalczykowski, Julio Fernandez, Matthias Rief, Herman Gaub, Sanford Leuba, Rich Superfine, Michelle Wang, and many others. I believe the labs I just mentioned perform research primarily in the "single-molecule biochemistry" quadrant. There are many examples of outstanding single-molecule analyses in the bottom-right quadrant. One of my recent favorites is from Osheim, Sikes, and Beyer where they used electron microscopy (EM) visualization of chromatin fibers extracted from Drosophila to study Pol II termination at the single-molecule level. "DNA fiber analysis" is another great example of research in this quadrant. The biomembrane force probe that Evan Evans' lab uses also lends itself to research that I would put in this quadrant. For example, the work of Heinrich, Leung, and Evans, studied ligand-receptor interactions on living human neutrophils. Another important example is from Cui and Bustamante, who studied the mechanical properties of individual native chromatin fibers from chicken erythrocytes.
The Cui and Bustamante work is closest to the killer application we are pursuing. We are working to use single-molecule DNA unzipping to map the positions of nucleosomes and polymerases on specific native chromatin fibers. This will be different from the Cui and Bustamante work in a number of ways. First, we expect to be able to map the positions of nucleosomes and polymerases with close to basepair accuracy. Second, we will analyze positions on site-specific chromatin fibers. Thus, we will know which gene it is, where the promoters and terminators are, etc. You can read more about our ideas in a recent minigrant proposal which I posted on Scribd. (This was funded by the way!) Our single-molecule research will complement the ensemble studies currently used -- commonly Chromatin Immunoprecipitation (ChIP). Because we will analyze chromatin extracted from living yeast cells (and higher organisms in the future), we will be able to study chromatin remodeling as genes are turned on or off and in any mutant strains we'd like. This is the same genetics as is carried out in the ensemble assays (top right quadrant), and in fact, our collaborators (Mary Ann Osley lab) currently do much research in this quadrant. We think our single-molecule method will be particularly good for addressing many open questions related to chromatin remodeling during transcription. For example, the Osley lab recently showed that a yeast double mutant deficient in FACT and H2b-ubiquitylation has an interesting phenotype that seems to have some kind of misassembled chromatin. Deciphering this chromatin structure is difficult with ensemble assays, and single-molecule analysis can shed a lot of light on this question. For example, if the chromatin is being assembled with histone tetramers instead of octamers, that should be clearly visible in the single-molecule unzipping signals.
By pursuing research in the "single-molecule genetics" or bottom-right quadrant, I think we are poised for making important contributions that complement the other quadrants. I just illustrated how we can closely complement ensemble genetics experiments (ChIP). Further, we complement single-molecule biochemstry experiments in the bottom-left and ensemble biochemistry in the top-left. As far as Pol II experiments go I don't believe any single-molecule force v. velocity transcription assays have been carried out yet. The work of Shundrovsky, Hall, and others in the Wang lab in terms of unzipping reconstituted mononucleosomes is in the bottom-left quadrant, and it certainly complements our work, because we're relying on their results to know what to look for when unzipping native chromatin fibers. And we'll do some of our own work in this quadrant in terms of unzipping Pol II in vitro transcription complexes for a similar purpose. In contrast to nucleosomes, we expect the Pol II unzipping signature to look distinct for unzipping from upstream versus downstream. If so, this will allow us to determine the sense versus antisense orientation of polymerases on native chromatin fibers, giving us single-molecule insight into a very exciting area of eukaryotic transcription. (See, for example, a recent antisense transcription paper from Core, Waterfall, and Lis.)
OK, hopefully I succeeded at least a bit in describing how I think we can lead a new area of single-molecule research: single-molecule genetics. I'd be curious in hearing whether this "2x2 matrix" helps you at all in looking at the research space. I find it very useful -- but on the other hand, I also really enjoyed a business course I took, and I thought the interview process for BCG was really fun. So, I may be a little different. If you do find it useful, I have some other 2x2's I can talk about. For example, Single-molecule/Ensemble versus With Force / Without Force.
*Footnote on "in vivo" terminology.
I am using "in vivo" to designate that the biology occurred in the context of a native cell, even if the analysis of the molecules was carried out with an in vitro assay. This would be true, in Chromatin Immunoprecipitation studies of chromatin remodeling during transcription, for example. The transcription is carried out in the nucleus, then the cells are fixed with formaldehyde and analysis is carried out in vitro. I am consdering this an "in vivo" experiment, in contrast to studies where the transription has been carried out in vitro -- such as in the amazing reconstituted systems of the Reinberg lab and others.
Certainly the holy grail of "in vivo" would be to know the 3-D position and chemical nature of every molecule in real-time while the cell is still living. And many single-molecule researchers are making big strides towards this goal by visualizing and tracking individual molecules inside living cells. However, my focus here is more on the arena of single-molecule manipulation.
**Footnote on doo doo
I may not be getting the of biochemistry and genetics exactly correct, and I do know that there is some ongoing rivalry between these two fields. John Lis, one of my science heroes who taught a molecular biology course I took told us two quotations. I wish I could remember who to attribute these to:
Famous geneticist: "Genetics without biochemistry is doo-doo."
Extremely measured response from famous biochemist: "Biochemistry without genetics is an exercise in frustration."
SJK Note 1: I don't even know enough to know whether I've remembered these correctly. Possibly interchange genetics<-->biochemistry, etc. but you get the point: they are complementary research fields and there is a rivalry.
SJK Note 2: If you know a source for these quotes, please post a comment! I couldn't find it on Google. Also see some interesting related discussion on friendfeed.

In this post, I want to elaborate on why I think our ideas are important but unique, and therefore not in direct competition with most of the leading labs in the single-molecule manipulation world today. The table below illustrates why I think we can lead a new era of experiments in an under-tapped area of single-molecule analysis--"single-molecule genetics." I am using a 2x2 matrix to analyze the research space. This is a technique I learned during the end of my graduate career when I interviewed and almost landed a job with Boston Consulting Group (BCG, a company I highly respected). The top row represents ensemble assays where the properties of many molecules are averaged together. The bottom row represents the single-molecule analysis research world. The left column represents experiments where the system being studied has been reconstituted from purified components. The right column represents experiments where the system being studied was intact, with only a few specific genes having been mutated or knocked-down*.

The top left quadrant represents what people refer to as biochemistry research, whereas the top right quadrant would be genetics**. The vast majority of the important discoveries in biology in the past half century would be in the top row (ensemble assays). Single-molecule assays in the bottom row have been very valuable, but are only now becoming more widely available. In my opinion, the vast majority of single-molecule assays have been in in the bottom left quadrant, which I am calling "single-molecule biochemistry" for convenience. For example, the Steve Block lab has made a number of breakthroughs in this quadrant. The Block Lab is a good example, because the research and papers produced by the students and postdocs in this lab over the past couple decades are of the highest quality and have played a key role in defining the single-molecule field. One of their areas of study is RNA Polymerase, where they have applied forces to recombinant E. coli RNA Polymerase during elongation in all sorts of manners: opposing elongation, assisting elongation, pulling on the RNA transcript, etc. Another main area of the block lab is the molecular motor kinesin. They have used optical tweezers to make many important discoveries about the mechanochemistry of this amazing enzyme. From these two areas combined (which still account for only part of the Block Lab research), they have something like 10,000 Science, Nature, and Cell papers. And as far as I know, all of that research has been done using dozens of different recombinant RNA Polymerase and kinesin heavy chain (KHC) motor proteins. So, as far as my matrix goes, they have made a huge impact on the bottom left quadrant -- "single molecule biochemsitry."
As I've said, I think the bottom right quadrant has so far been under-utilized. I am calling this quadrant "single-molecule genetics," to highlight what I see as the untapped power of combining proven single-molecule analyses with existing genetics techniques. When I say "untapped," I don't mean to imply that there aren't existing studies in this quadrant. I only mean that most of the research (and money) from single-molecule manipulation groups I am familiar with has been going into the SM biochemistry quadrant. This includes groups such as those of Steve Block, Carlos Bustamante, Evan Evans, Mark Williams, Jeff Gelles, Vincent Croquette, David Bensimon, Steve Kowalczykowski, Julio Fernandez, Matthias Rief, Herman Gaub, Sanford Leuba, Rich Superfine, Michelle Wang, and many others. I believe the labs I just mentioned perform research primarily in the "single-molecule biochemistry" quadrant. There are many examples of outstanding single-molecule analyses in the bottom-right quadrant. One of my recent favorites is from Osheim, Sikes, and Beyer where they used electron microscopy (EM) visualization of chromatin fibers extracted from Drosophila to study Pol II termination at the single-molecule level. "DNA fiber analysis" is another great example of research in this quadrant. The biomembrane force probe that Evan Evans' lab uses also lends itself to research that I would put in this quadrant. For example, the work of Heinrich, Leung, and Evans, studied ligand-receptor interactions on living human neutrophils. Another important example is from Cui and Bustamante, who studied the mechanical properties of individual native chromatin fibers from chicken erythrocytes.
The Cui and Bustamante work is closest to the killer application we are pursuing. We are working to use single-molecule DNA unzipping to map the positions of nucleosomes and polymerases on specific native chromatin fibers. This will be different from the Cui and Bustamante work in a number of ways. First, we expect to be able to map the positions of nucleosomes and polymerases with close to basepair accuracy. Second, we will analyze positions on site-specific chromatin fibers. Thus, we will know which gene it is, where the promoters and terminators are, etc. You can read more about our ideas in a recent minigrant proposal which I posted on Scribd. (This was funded by the way!) Our single-molecule research will complement the ensemble studies currently used -- commonly Chromatin Immunoprecipitation (ChIP). Because we will analyze chromatin extracted from living yeast cells (and higher organisms in the future), we will be able to study chromatin remodeling as genes are turned on or off and in any mutant strains we'd like. This is the same genetics as is carried out in the ensemble assays (top right quadrant), and in fact, our collaborators (Mary Ann Osley lab) currently do much research in this quadrant. We think our single-molecule method will be particularly good for addressing many open questions related to chromatin remodeling during transcription. For example, the Osley lab recently showed that a yeast double mutant deficient in FACT and H2b-ubiquitylation has an interesting phenotype that seems to have some kind of misassembled chromatin. Deciphering this chromatin structure is difficult with ensemble assays, and single-molecule analysis can shed a lot of light on this question. For example, if the chromatin is being assembled with histone tetramers instead of octamers, that should be clearly visible in the single-molecule unzipping signals.
By pursuing research in the "single-molecule genetics" or bottom-right quadrant, I think we are poised for making important contributions that complement the other quadrants. I just illustrated how we can closely complement ensemble genetics experiments (ChIP). Further, we complement single-molecule biochemstry experiments in the bottom-left and ensemble biochemistry in the top-left. As far as Pol II experiments go I don't believe any single-molecule force v. velocity transcription assays have been carried out yet. The work of Shundrovsky, Hall, and others in the Wang lab in terms of unzipping reconstituted mononucleosomes is in the bottom-left quadrant, and it certainly complements our work, because we're relying on their results to know what to look for when unzipping native chromatin fibers. And we'll do some of our own work in this quadrant in terms of unzipping Pol II in vitro transcription complexes for a similar purpose. In contrast to nucleosomes, we expect the Pol II unzipping signature to look distinct for unzipping from upstream versus downstream. If so, this will allow us to determine the sense versus antisense orientation of polymerases on native chromatin fibers, giving us single-molecule insight into a very exciting area of eukaryotic transcription. (See, for example, a recent antisense transcription paper from Core, Waterfall, and Lis.)
OK, hopefully I succeeded at least a bit in describing how I think we can lead a new area of single-molecule research: single-molecule genetics. I'd be curious in hearing whether this "2x2 matrix" helps you at all in looking at the research space. I find it very useful -- but on the other hand, I also really enjoyed a business course I took, and I thought the interview process for BCG was really fun. So, I may be a little different. If you do find it useful, I have some other 2x2's I can talk about. For example, Single-molecule/Ensemble versus With Force / Without Force.
*Footnote on "in vivo" terminology.
I am using "in vivo" to designate that the biology occurred in the context of a native cell, even if the analysis of the molecules was carried out with an in vitro assay. This would be true, in Chromatin Immunoprecipitation studies of chromatin remodeling during transcription, for example. The transcription is carried out in the nucleus, then the cells are fixed with formaldehyde and analysis is carried out in vitro. I am consdering this an "in vivo" experiment, in contrast to studies where the transription has been carried out in vitro -- such as in the amazing reconstituted systems of the Reinberg lab and others.
Certainly the holy grail of "in vivo" would be to know the 3-D position and chemical nature of every molecule in real-time while the cell is still living. And many single-molecule researchers are making big strides towards this goal by visualizing and tracking individual molecules inside living cells. However, my focus here is more on the arena of single-molecule manipulation.
**Footnote on doo doo
I may not be getting the of biochemistry and genetics exactly correct, and I do know that there is some ongoing rivalry between these two fields. John Lis, one of my science heroes who taught a molecular biology course I took told us two quotations. I wish I could remember who to attribute these to:
Famous geneticist: "Genetics without biochemistry is doo-doo."
Extremely measured response from famous biochemist: "Biochemistry without genetics is an exercise in frustration."
SJK Note 1: I don't even know enough to know whether I've remembered these correctly. Possibly interchange genetics<-->biochemistry, etc. but you get the point: they are complementary research fields and there is a rivalry.
SJK Note 2: If you know a source for these quotes, please post a comment! I couldn't find it on Google. Also see some interesting related discussion on friendfeed.




